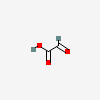|
|
|
| Reference: |
Meyer P, Liger D, Leulliot N, Quevillon-Cheruel S, Zhou CZ, Borel F, Ferrer JL, Poupon A, Janin J, van Tilbeurgh HCrystal structure and confirmation of the alanine:glyoxylate aminotransferase activity of the YFL030w yeast proteinBiochimie v87, p.1041-1047We have determined the three-dimensional crystal structure of the protein encoded by the open reading frame YFL030w from Saccharomyces cerevisiae to a resolution of 2.6 A using single wavelength anomalous diffraction. YFL030w is a 385 amino-acid protein with sequence similarity to the aminotransferase family. The structure of the protein reveals a homodimer adopting the fold-type I of pyridoxal 5"-phosphate (PLP)-dependent aminotransferases....
|
| Description: |
Yeast Alanine:glyoxylate Aminotransferase Yfl030w. |
| Deposition: |
2005/2/21   |
| |
| Author: |
Meyer P, Liger D, Leulliot N, Quevillon-Cheruel S, Zhou CZ, Borel F, Ferrer JL, Poupon A, Janin J, Van Tilbeurgh H |
|
| Taxonomy: |
Saccharomyces cerevisiae |
| Related Structure: |
VAST |
|
|
Molecular components in the MMDB structure are listed below and may include macromolecular chains, 3D domains, protein classifications (domain families), and ligands, as available. Mouse over each icon for more information on the component.  | Ligand |
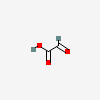 |
 |
glyoxylic acid |
|
1 occurrence |
|
|
Citing MMDB:
 Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, Zhang N, Bryant SH.
"MMDB: annotating protein sequences with Entrez's 3D-structure database.",
Nucleic Acids Res. 2007 Jan; 35(Database issue): D298-300.
Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, Zhang N, Bryant SH.
"MMDB: annotating protein sequences with Entrez's 3D-structure database.",
Nucleic Acids Res. 2007 Jan; 35(Database issue): D298-300.
 Chen J, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler-Bauer A, Marchler GH, Mazumder R, Nikolskaya AN, Rao BS, Panchenko AR, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH.
"MMDB: Entrez's 3D-structure database", Nucleic Acids Res. 2003 Jan; 31(1): 474-7.
Chen J, Anderson JB, DeWeese-Scott C, Fedorova ND, Geer LY, He S, Hurwitz DI, Jackson JD, Jacobs AR, Lanczycki CJ, Liebert CA, Liu C, Madej T, Marchler-Bauer A, Marchler GH, Mazumder R, Nikolskaya AN, Rao BS, Panchenko AR, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Vasudevan S, Wang Y, Yamashita RA, Yin JJ, Bryant SH.
"MMDB: Entrez's 3D-structure database", Nucleic Acids Res. 2003 Jan; 31(1): 474-7.
|
 |