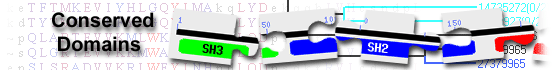?
Short-chain alcohol dehydrogenase. This family contains a number of bacterial short-chain alcohol dehydrogenases that are approximately 400 residues long. Alcohol dehydrogenases display a wide variety of substrate specificities, and play an important role in a broad range of physiological processes. Short-chain alcohol dehydrogenases form part of a group of alcohol dehydrogenases that are dependent upon NADP.