Research Highlights
September 2008
New Data Analysis Tools Advance Protein Research
PNNL provides no-cost resources to scientists around the world
Results: From developing viable bioenergy to detecting disease, proteins are crucial structural and functional elements to all biological functions. Proteomics experts from Pacific Northwest National Laboratory and two universities have collaborated to develop and deploy data analysis tools to further the field of protein research. Two of these tools are now available free of charge through publicly available websites.
DAnTE (Data Analysis Tool Extension) was developed as a statistical and visualization software that scientists can use to perform data analysis steps on large scale proteomics data. These include normalization algorithms that can correct systematic variations, algorithms that can estimate and impute missing values—a major issue associated with proteomics data—peptide to protein rollup algorithms, and hypothesis testing methods. Though designed specifically for analyzing proteomics data, DAnTE performs equally well on genomics microarray data. The software can be downloaded at http://omics.pnl.gov/.
The second tool now available is a data analysis strategy for temporal "bottom-up" proteomics data obtained using mass spectrometry methods that can detect thousands of peptides over time. The strategy uses algorithms to normalize protein abundance, impute missing values, and infer dynamic patterns of peptides and proteins. This framework was demonstrated on data from a time-course study on Rhodobacter sphaeroides that examined the transition between aerobic respiration and photosynthesis. R. sphaeroides is an environmentally important photosynthetic microbe that grows under a variety of conditions using a variety of electron acceptors. The code is publicly available at http://ober-proteomics.pnl.gov/software/.
Why it matters: Better tools for protein identification are vital to solving intractable problems such as converting agricultural waste into fuels, detecting bio-based threats and quickly detecting and treating disease. Making new proteomics tools available at no cost to the scientific community allows more researchers to enter the proteomics field without investing in expensive tools or needing to develop their own.
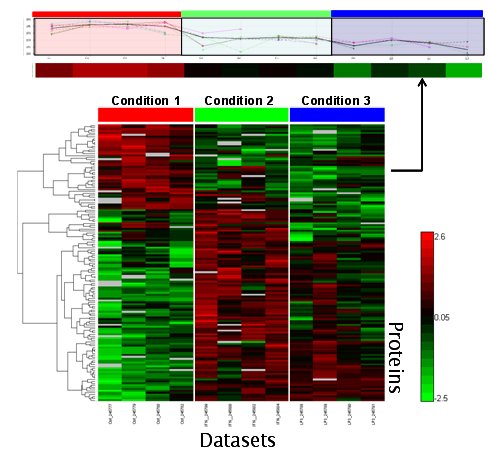 Plot from DAnTE shows a candidate set of proteins identified as significant biomarkers that differentiate three treatment conditions. Top bar shows the profile of one protein from the set.
Plot from DAnTE shows a candidate set of proteins identified as significant biomarkers that differentiate three treatment conditions. Top bar shows the profile of one protein from the set.
Acknowledgments: The DAnTE development team includes Ashoka Polpitiya, Wei-Jun Qian, Navdeep Jaitly, Vladislav Petyuk, Joshua Adkins, David Camp, Gordon Anderson and Richard Smith. The research team for the temporal data analysis framework includes former PNNL scientist Xiuxia Du, Stephen Callister, Nathan Manes, Joshua Adkins, Richard Smith and Mary Lipton, all PNNL; Roxana Alexandridis, Xiaohua Zeng, Jung Hyeob Roh, William Smith and Samuel Kaplan, University of Texas; and Timothy Donohue, University of Wisconsin-Madison.
The DAnTE research was funded by the National Institutes of Health's National Institute of General Medical Sciences, National Center for Research Resources, the National Institute of Allergy and Infectious Diseases and PNNL's Next Generation Proteomics Measurement Platform. The data analysis framework research was supported by DOE's Office of Biological and Environmental Research and NIH's NIAID and National Institute of General Medical Sciences. Portions of the research for both tools were performed in the Environmental Molecular Sciences Laboratory, a DOE-BER national scientific user facility located at PNNL.
References: Du X, SJ Callister, NP Manes, JN Adkins, RA Alexandridis, X Zeng, JH Roh, WE Smith, TJ Donohue, S Kaplan, RD Smith, and MS Lipton. 2008. "A Computational Strategy to Analyze Label-Free Temporal Bottom-Up Proteomics Data." Journal of Proteome Research 7(7):2595-604, July 2008.
Polpitiya AD, WJ Qian, N Jaitly, VA Petyuk, JN Adkins, DG Camp II, GA Anderson, and RD Smith. 2008. "DAnTE: A Statistical Tool for Quantitative Analysis of -omics Data." Bioinformatics 24(13):1556-8, July 2008.

